Results and discussion
Results
Test Case
Let’s consider the case of a forest in Kornanwand (Bas-Rhin, France) located at the West of Haguenau. This is a small forest amid fields.
The Location is define by it bounding box following the Well-Known Text (WKT) Standard (ISO/IEC 13249-3:2016). The coordinates are given in Lambert93 the standard Coordinate Reference System (CRS) in France.
On the map, the shape of the forest is represented as defined in BD TOPO.
Starting Point

Before my intership the forest were represented only by their canopy (figure 1). Using some of the algorithms describe above we will first detect the position of the tree using a local maximum filter then populatethe forest with trees generated by the method of Weber and Penn.
Detection
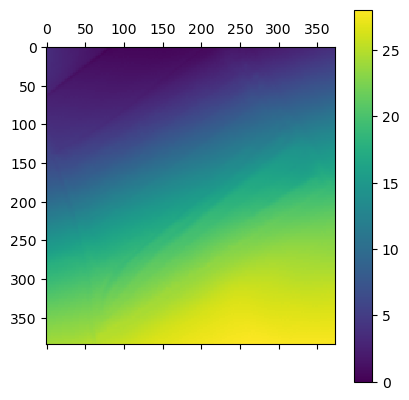
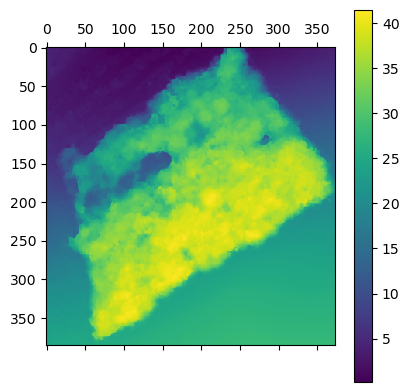
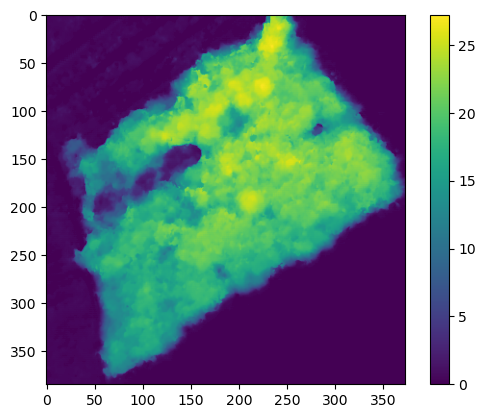
Th first Step in the detection of trees is to build a CHM. With the code I previously obtain I get the DTM and DSM the given zone. Then I computed the difference to get the height of the canopy (CHM)
To detect tree position I simply used a local maximum filter with the following parameters
- min_distance
-
minimum number of space between to tree.
- threshold_abs
-
minum size in meter to be classified as a tree.
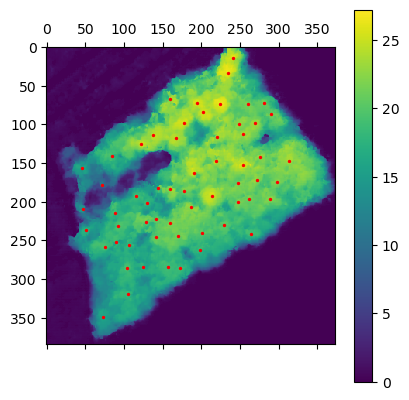
min_dist=10, threshold_abs=5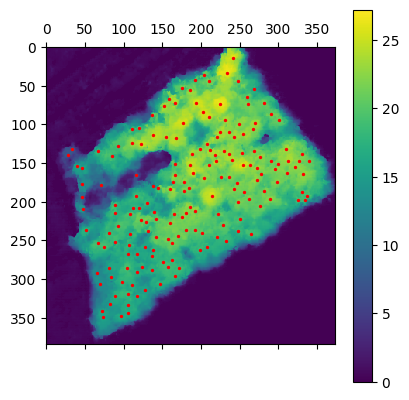
min_dist=5, threshold_abs=5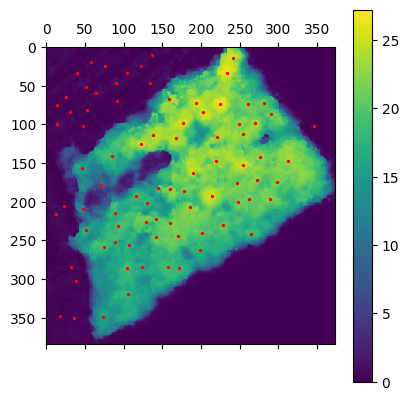
min_dist=10, threshold_abs=0One can observe that the both parameter have a massive impact on the number false positive of false negative. In fact in min_distance is to high we miss trees and is threshold is to low we could
classify ground point as trees.
Results
Combining the two results previously obtained I compute the discrete representation of the forest.
I ran the generation on a laptop with a quad-core CPU (Intel® Core™ i5-7300HQ CPU @ 2.50GHz)
and 8 Gb of RAM.
The program was parallelized using the python multiprocessing library. It took 20 minutes to generate 150 trees. Each tree is generate on its own file in the .ply format.
In figure 1O, I displayed only a part of the forest, because my ram doesn’t allow me to display more.
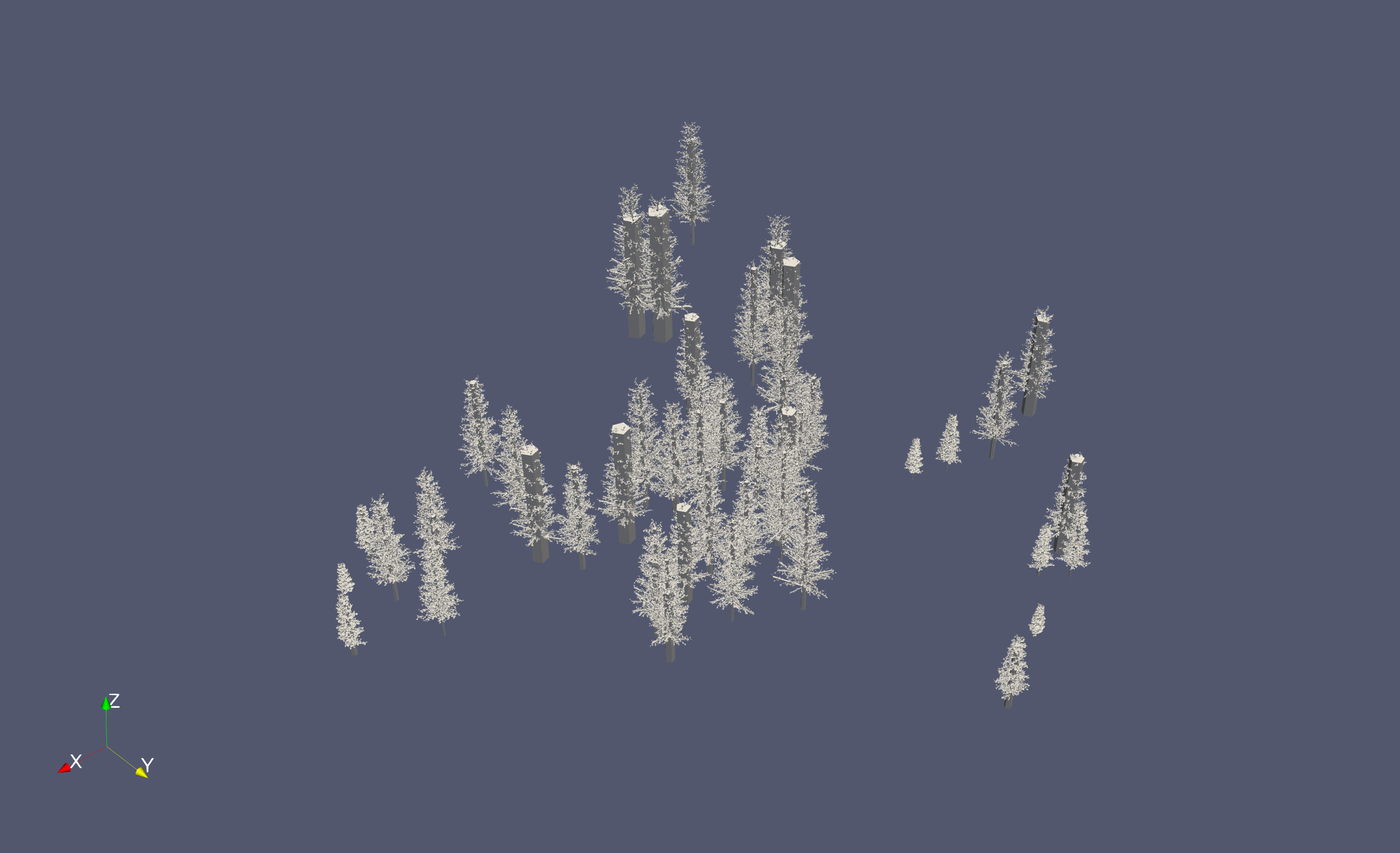
Improvements
The tool I developed still requires some improvements.
Hybrid Method
First the use of an hybrid method between using Weber and Penn to initialize the space colonization algorithm would be a good compromise to speed up and fit more closely to the DSM. Though this hybrid is in development (figure 11).
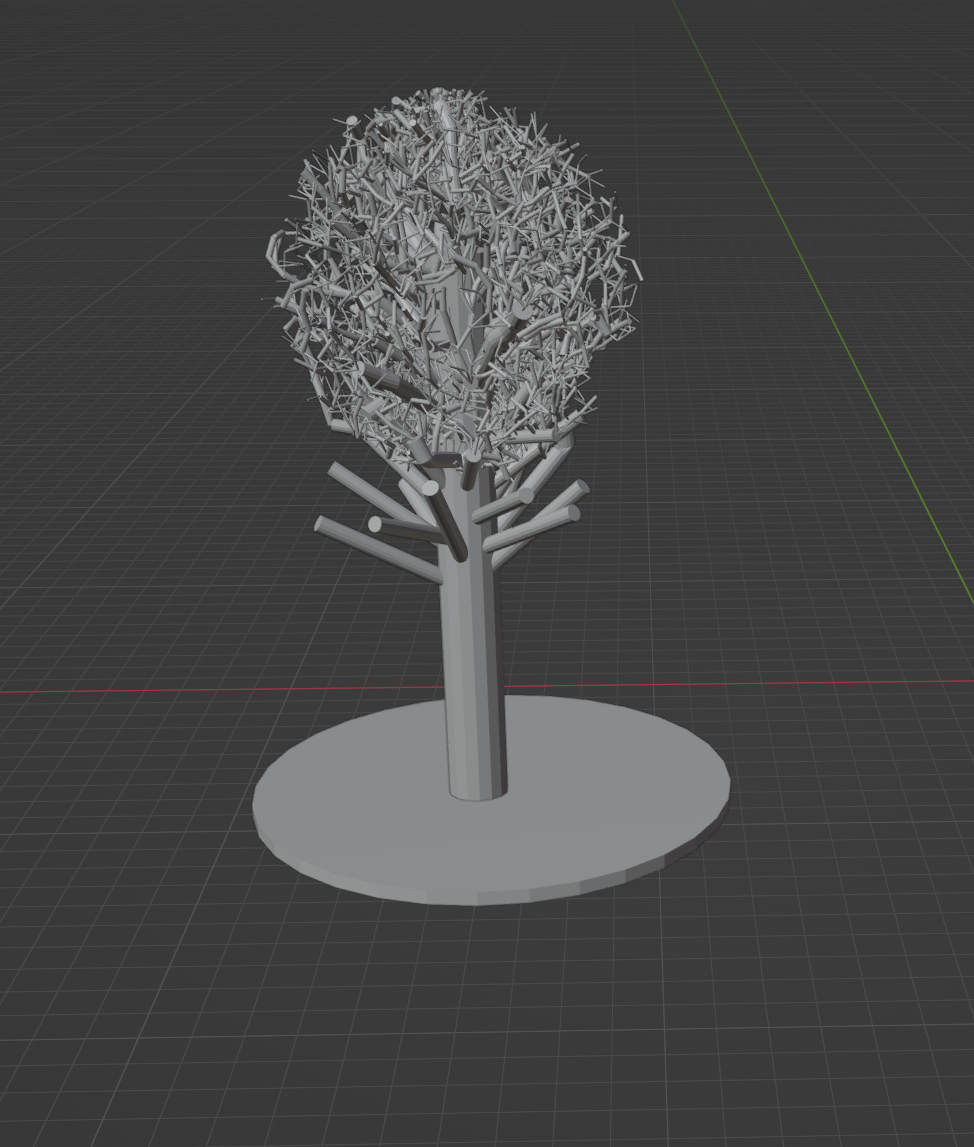
Detection
To be applying the space colonization algorithm needs to be feed attraction point, to do so segmentation algorithm like Dalponte or Silva should be applied.
Performance
The implementation was made using python, this could be speed up by integating some C++ and by using MPI for the parallelization. Each beeing contruct independantly from it’s neighbourgs, it will be easy to integrate MPI to the code as it would require few communication between the threads.
 .pdf
.pdf

